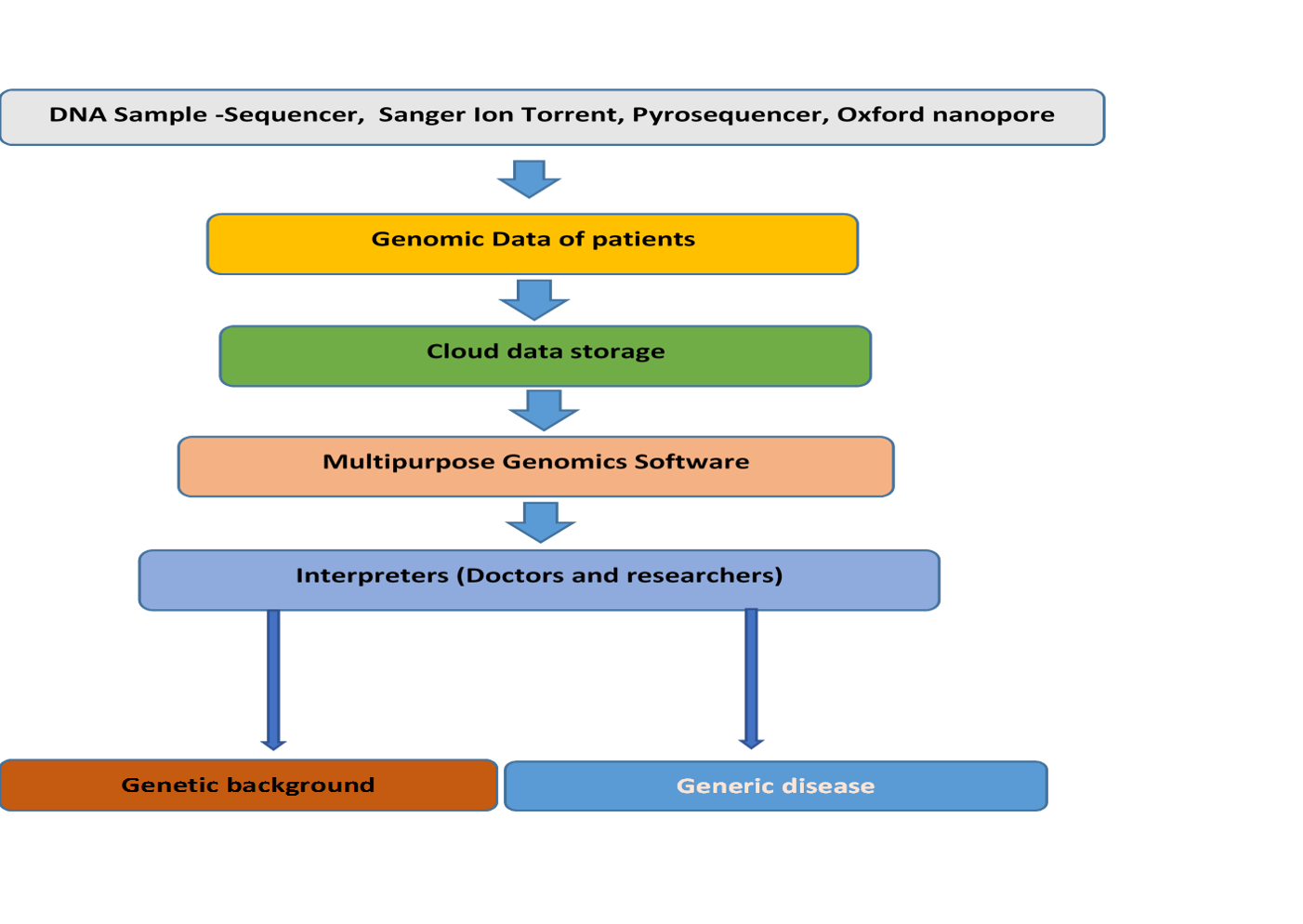
- We use Machine learning applications for the analysis of genome sequencing data sets. The technology used in this software will be able to read, Trim, assemble, annotate, and process different sequencer output data, including FASTA, FASTQ, and FAST5, and finally, will analyze the raw data with the genebanks to produce applicable information, including the name of the genes, and the presence of any abnormalities, which finally illustrates the genetic status of the case. Deep learning would be used to find patterns in the data. A Deep Neural Network (DNN) would be located on the cloud, and sequenced human genomes from our client database would be uploaded. With time being of the essence, parallel computing will reduce the time required for the DNN (RNN ) to perform its operation in a matter of hours. ML is performed in five phases.
- 1) Algorithm selection.
- 2) Data extraction, transformation, and loading where the data is formatted and then cleaned to get rid of imperfect data and attend to missing data. The data may also be transformed by scaling, decomposition, or aggregation.
- 3) The data are split into training and testing sets. The training set is used to develop a model or “train” the artificial intelligence (AI). The parameters in the model may be modified to enhance the final model
- 4) The model is used to make predictions on the testing set, and the predictability of the model is assessed. Cross-validation can be performed to compare the predicted outcome to the actual patient outcome. Sensitivity, accuracy, and receiver operating characteristic (ROC) curves may also be used to evaluate the predictability of the model.
- 5) The application is implemented to make predictions for other data sets.
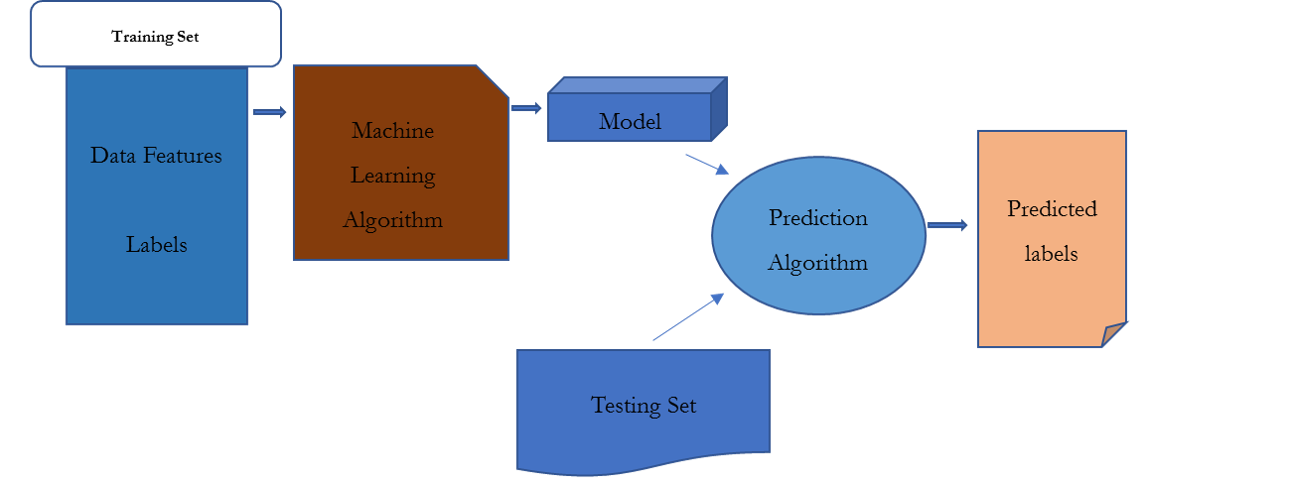
Machine Learning Model Creation
A training set of DNA sequences is supplied as input to a learning procedure, along with binary labels indicating whether each sequence is centered. The learning algorithm delivers a model that can then be consequently applied, in conjunction with a prediction algorithm, to assign predicted labels to unlabeled test sequences