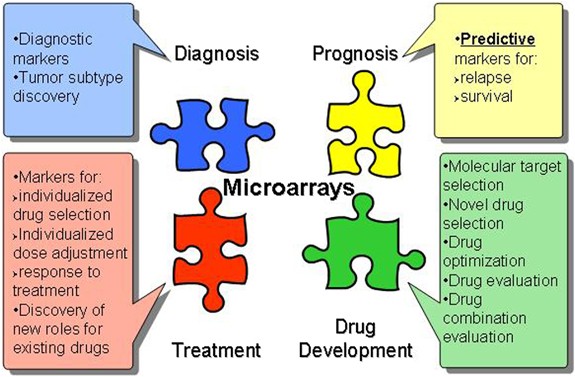
Pharmacogenomics is the study of genetic variation that is associated with the variable responses of individuals to any given drug treatment , including individual differences in drug efficacy and susceptibility to adverse effects. This area of genomics provides possibly the best and clearest example of how genomics can be used to bring about more targeted and individualized treatments and actually influence clinical care. This area has already made a number of significant impacts in this regard. Specifically, over the past several years, many associations between genetic variants and drug response have been discovered, including, for example, the now well-known association between CYP2C9 and VKORC1 gene variants and Warfarin .
Warfarin is one of the most commonly used anticoagulant medications and is prescribed worldwide to prevent stroke and venous thromboembolism . Unfortunately, however, dosing of Warfarin is highly complex due to many factors that affect its metabolism, including clinical (drug-drug interactions, dietary interactions, age, and body surface area) and genetic factors, in particular variants in the genes CYP2C9 and VKORC1 . In fact, it is estimated that consideration of combined genotypes across variants in these genes, together with factors such as age and body size, are estimated to account for 35–60% of the variability in Warfarin dosing requirements. Evidence for the importance of these variants in influencing Warfarin metabolism led the U.S. Food and Drug Administration (FDA) to update the labeling of Warfarin in 2007 to include a statement acknowledging the importance and potential of genotyping during the early phase of dosing . This particular update, however, did not dictate how physicians should change the dosage based on genotype. Based on additional research since then, the FDA again updated its Warfarin labeling in 2010 to include specific ranges of initial doses assigned to each genotype representing the expected steady state maintenance doses.
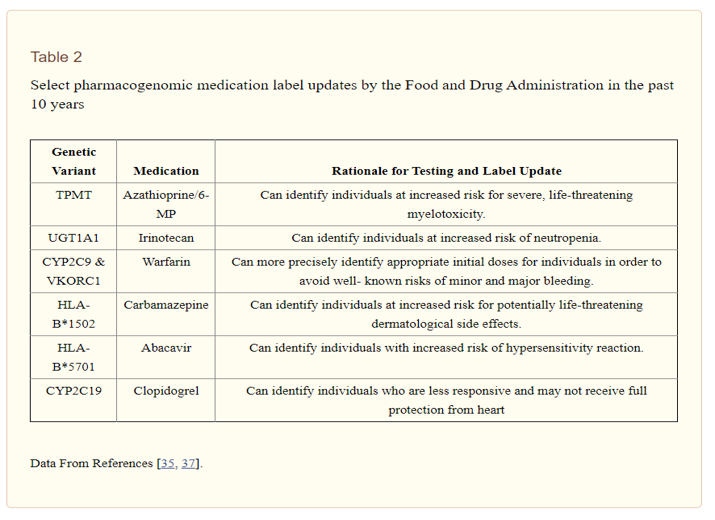
To the extent that pharmacogenomic research efforts and resulting label updates by the FDA lead to improved drug safety and efficacy, this represents a real step forward in the use of genomics for disease treatment (and prevention of adverse effects) and for ushering in a new era of personalized medicine. Other examples of pharmacogenomic associations and FDA label updates are included in Table 2 . Briefly, Azathioprine and 6-MP are immunosuppressants used to treat some cancers, as well as autoimmune diseases such as rheumatoid arthritis and Crohn’s disease. Genetic testing for variants in the TPMT gene have been found to be associated with adverse side effects, including severe myelotoxicity, and the 6-MP label was the first label to be updated in the last decade based on pharmacogenomic information. A similar association was found between Irinotecan, an anti-cancer chemotherapy drug, and variations in the gene UGT1A1. This was the first label update to recommend a specific dosing reduction based on pharmacogenomics because of an increased risk of neutropenia in patients with certain genotypes. In terms of mental health and applications to neurological disorders, Carbamazepine, an anticonvulsant and mood stabilizer used in the treatment of epilepsy, bipolar disorder, trigeminal neuralgia, and other neuropsychiatric disorders, has shown an association with variants in HLA-B*1502. Specifically, genotyping of variants in HLA-B*1502 can identify individuals at risk for Stevens-Johnson syndrome and toxic epidermal necrolysis, two forms of rare but potentially fatal skin diseases . Similarly, associations between Abacavir and variants in HLA-B*5701, have also been shown to predict adverse drug effects. Finally, another notable pharmacogenomic association is that between genetic variants in CYP2C19 and metabolism of Clopidogrel, which is used in at-risk patients to prevent strokes and heart attacks. In 2010, the label of Clopidogrel was updated with a warning that patients carrying variants associated with poor metabolism of the drug may be less responsive to the medication and thus fail to receive full protection from heart attacks, stroke, and cardiovascular death .
While pharmacogenomic research and findings have led the FDA to take major steps forward in translating genomic findings into better, individualized disease treatments, there is still much work to be done. For example, with respect to pharmacogenomics in psychiatry, limited evidence from twin and family studies suggests that response to antipsychotic and antidepressant medications are heritable traits. Specifically, studies on single pairs of monozygotic twins observed similar response to treatment with antipsychotics and similar levels of drug-induced weight gain , and studies on siblings and first-degree relatives observed similarities in treatment-induced tardive dyskinesia and response to antidepressants . Furthermore, genetic variants in the HLA complex have been associated with risk of drug-induced agranulocytosis, which led a biotechnology company to develop and offer a genetic test for the determination of high (1.5%) or low (0.5%) risk in conjunction with prescription of clozapine . This test, however, has not been widely adopted by physicians , and the FDA has not updated labels of major antidepressants or antipsychotics, in part, due to difficulties replicating findings in this area and a lack of large-scale controlled studies. Another issue that may inhibit translation of pharmacogenomic findings into the clinic is a lack of adequate training of community physicians in pharmacogenomics. Furthermore, the clinical availability of genetic testing for pharmacogenomic variants is still limited in many areas, and there remain important questions with respect to insurance reimbursement and who will pay for testing.
Although not a pharmacogenomic association per se, another genetic association that may have implications for the treatment of unipolar depression is the well-studied gene-environment interaction between the promoter region of the serotonin transporter gene (5-HTTLPR) and stressful life events. In a large cohort, childhood abuse and stressful life events were associated with a high risk of becoming depressed in individuals with the short allele of the 5-HTTLPR, but had little effect on the development of depression among long allele homozygotes . Further work stemming from this initial report suggests that the finding has more general implications in that this variant is likely not directly associated with depression, but that the 5-HTTLPR, together with polymorphisms in other genes such as BDNF and CRHR1 , are more broadly associated with personal dispositions that are more or less sensitive to environmental surroundings. In terms of treatment for depression, psychological therapy and antidepressant medications have, on average, comparable efficacy in unselected groups of patients diagnosed with depression. Importantly, however, the potential gene-environment interaction involving 5-HTTLPR suggests that individuals who are more sensitive to environmental stimuli may respond better to psychological treatments versus antidepressant medication . Although this particular gene-environment interaction has been called into question in recent years , these findings illustrate the potential importance of further study of gene-environment interactions in other contexts, as well as the potential implications of such findings for disease treatment and prevention in psychiatry. Another issue particularly relevant for mental health conditions such as depression is that of placebo effects and the cyclical nature of the conditions themselves, which both have implications for drug and pharmacogenomic discovery in this particular disease area.
