It is important to note that genetics and genomics are complementary but different disciplines. While genetics is the study of inheritance, or the way traits are passed down from one generation to another, genomics refers to the study of all the genes and gene products in an individual, as well as how those genes interact with one another and the environment. In 1866, Gregor Mendel published his theories of inheritance based on hybridization experiments in garden peas. Although largely ignored until the early 1900’s, his work is considered to form the basic theory of modern genetics. Mendel discovered that when he crossed a white flower and a purple flower plant, rather than being a mix of the two, the offspring were purple flowered. From several successive crosses, he developed the ideas of dominant and recessive inheritance in which dominant factors (purple flowered) will hide recessive factors (white flowered). Later, in the early 1900s, Sir Archibald Garrod described several human diseases that he termed “inborn errors of metabolism” (e.g., albinism), which showed a similar pattern of inheritance to that of the white color in Mendel’s hybridization experiments and thus suggested a genetic basis for these diseases . Garrod described the nature of autosomal recessive inheritance (where two copies of an abnormal genetic variant or “allele” are needed to express the disease) with respect to these conditions, and using Mendelian principles illustrated that two parents heterozygous for the disease allele (i.e., who carry only one copy) have a one-in-four chance of producing a child having both disease alleles and, hence, the unwanted disorder.

In 1909 Wilhelm Johannsen coined the term “gene”, and in 1915, Thomas Morgan Hunt showed that genes, which exist on chromosomes, are the basic units of inheritance . Over the next several years, many additional examples of diseases inherited in an autosomal recessive manner were discovered, including phenylketonuria and cystic fibrosis. Autosomal dominant diseases (where only one copy of an abnormal allele is needed to express the disease) were also described, including Huntington’s disease and Marfan’s syndrome. X-linked recessive and X-linked dominant traits (see Glossary of Terms) were also discovered and described. Importantly, disorders that follow these patterns of inheritance, termed “Mendelian disorders”, have been among the easiest to analyze and the best understood. Arguably, these disorders and modes of inheritance also have led to a common fatalistic view among lay people that genes are deterministic and that if one inherits a disease allele, one will inherit an unwanted disorder. This view stands in opposition to more recent notions of the “susceptibility allele”, based in large part on findings from the field of genomics, specifically GWAS, in which inheritance of a specific genetic variant does not guarantee the emergence of a disease, but merely confers some environmentally and/or other genetic factor mediated probability of developing a disease .
In 1920 Hans Winkler coined the term “genome” to refer to all the genes in an organism by combining the words gene and chromosome . Building on evidence that DNA, or deoxyribonucleic acid, was the primary component of chromosomes and genes, in 1953 James Watson and Francis Crick, with the help of Rosalind Franklin, discovered the double helix structure of DNA, and in 1966 Marshall Nirenberg was credited with “cracking the genetic code” and describing how it operates to make proteins . From these important discoveries, several downstream events occurred that began to reflect progression from the study of inheritance to the study of how genomic profiles could be used in medicine and in other arenas. For instance, in 1978, the biotechnology company Genentech, Inc. genetically engineered bacteria to produce human insulin (the first drug made through the use of recombinant DNA technology) for the treatment of diabetes , and in 1983 scientists identified the gene responsible for Huntington’s disease, which led to the first genetic test for a disease. In 1984, Sir Alec John Jeffreys developed DNA profiling or “fingerprinting” to be used in paternity testing and forensics, and in 1986, Richard Buckland was the first person acquitted of a crime based on DNA evidence. In 1989, Stephen Fodor, who later co-founded the gene chip company Affymetrix, developed the first DNA “microarray” and scanner, which would eventually lead to a method for testing hundreds of thousands of genetic variants simultaneously and thus foreshadowing the upcoming era of genome-wide association studies (GWAS) .
More recently, two major research initiatives have led to the present day “post-genomics” era – i.e., the Human Genome Project (HGP) and the International HapMap Project. The goal of the HGP was to draft a reference human genome delineating the sequence of chemical base pairs which make up DNA, as well as to determine the location of the roughly 25,000 genes thought to populate the human genome. In 2000 a draft sequence was released and in 2003 the final version was published. Relatedly, the goal of the International HapMap Project was to create a genome-wide database cataloging patterns of common human sequence variation within and across both individuals and populations. Importantly, an explicit aim of the HapMap was to facilitate identification of commonly occurring complex (i.e., non-Mendelian) disease causing genetic variants based on the “common disease, common variant” hypothesis , a hypothesis that incidentally has been called into question for explaining the genetic contributions to mental illness . This hypothesis suggests that genetic influences on complex diseases are attributable to common allelic variants present in more than 5% of the population. These variants represent “risk factors” or “susceptibility variants” for disease, as opposed to the more deterministic variants governed by Mendel’s principles. Taken together with technological developments enabling cost effective applications of DNA microarrays capable of measuring hundreds of thousands of genetic variants at once, these two initiatives led to the first of many GWAS to be published. While GWAS have some weaknesses, which will be discussed in later sections of this article, it is this research paradigm (now together with whole-genome sequencing) that has generally been thought of as laying the groundwork for the era of genomic and personalized medicine.
Heritability of Major Chronic Diseases
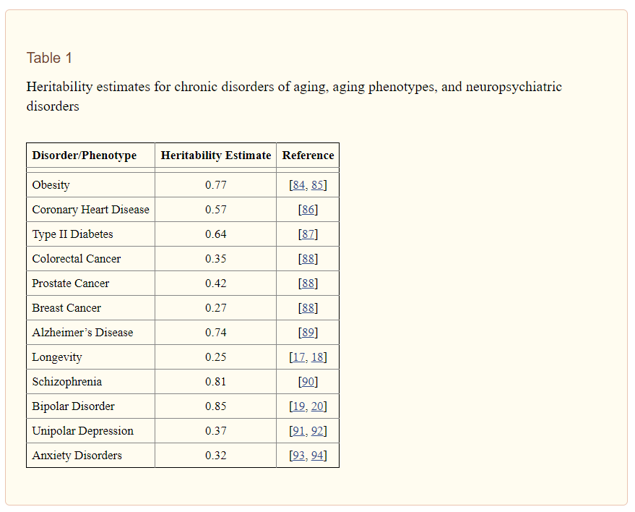
It is important to make explicit the reasons for considering genetics and genomics to have substantial utility for disease treatment and prevention, especially given the obvious role of environmental (including behavioral) factors in the etiology of many conditions (e.g., the role of smoking in the development of lung cancer), including neuropsychiatric disorders. Essentially, from family, twin, and adoption studies, researchers have been able to estimate the proportion of variance due to genetics (i.e., “heritability”) and the proportion of variance due to environment for a range of chronic diseases and other phenotypes. A heritability estimate of 1.0 indicates that all of the variation in a trait can be accounted for by genetics, and a heritability estimate of 0 indicates that all of the variation in a trait can be accounted for non-genetic factors (e.g., environment). Table 1 gives heritability estimates for chronic disorders of aging, aging phenotypes, and major neuropsychiatric disorders. These estimates range from 0.25 for human longevity to 0.85 for Bipolar disorder , indicating a strong genetic component with respect to many chronic diseases and phenotypes relevant for human health, including mental health. Ultimately, the large extent to which human disease can be attributable to genetic factors underscores the importance of genetics and genomics research for impacting health care.